BaseUnfold
- class deepinv.unfolded.BaseUnfold(iterator, params_algo={'lambda': 1.0, 'stepsize': 1.0}, data_fidelity=None, prior=None, max_iter=5, trainable_params=['lambda', 'stepsize'], device=device(type='cpu'), *args, **kwargs)[source]
Bases:
BaseOptimBase class for unfolded algorithms. Child of
deepinv.optim.BaseOptim.Enables to turn any iterative optimization algorithm into an unfolded algorithm, i.e. an algorithm that can be trained end-to-end, with learnable parameters. Recall that the algorithms have the following form (see
deepinv.optim.OptimIterator()):\[\begin{split}\begin{aligned} z_{k+1} &= \operatorname{step}_f(x_k, z_k, y, A, \gamma, ...)\\ x_{k+1} &= \operatorname{step}_g(x_k, z_k, y, A, \lambda, \sigma, ...) \end{aligned}\end{split}\]where \(\operatorname{step}_f\) and \(\operatorname{step}_g\) are learnable modules. These modules encompass trainable parameters of the algorithm (e.g. stepsize \(\gamma\), regularization parameter \(\lambda\), prior parameter (g_param) \(\sigma\) …) as well as trainable priors (e.g. a deep denoiser).
- Parameters:
iteration (str, deepinv.optim.OptimIterator) – either the name of the algorithm to be used, or directly an optim iterator. If an algorithm name (string), should be either
"GD"(gradient descent),"PGD"(proximal gradient descent),"ADMM"(ADMM),"HQS"(half-quadratic splitting),"CP"(Chambolle-Pock) or"DRS"(Douglas Rachford). See <optim> for more details.params_algo (dict) – dictionary containing all the relevant parameters for running the algorithm, e.g. the stepsize, regularisation parameter, denoising standard deviation. Each value of the dictionary can be either Iterable (distinct value for each iteration) or a single float (same value for each iteration). Default:
{"stepsize": 1.0, "lambda": 1.0}. See Parameters for more details.deepinv.optim.DataFidelity (list,) – data-fidelity term. Either a single instance (same data-fidelity for each iteration) or a list of instances of
deepinv.optim.DataFidelity()(distinct data-fidelity for each iteration). Default:None.prior (list, deepinv.optim.Prior) – regularization prior. Either a single instance (same prior for each iteration) or a list of instances of deepinv.optim.Prior (distinct prior for each iteration). Default:
None.max_iter (int) – number of iterations of the unfolded algorithm. Default: 5.
trainable_params (list) – List of parameters to be trained. Each parameter should be a key of the
params_algodictionary for thedeepinv.optim.OptimIterator()class. This does not encompass the trainable weights of the prior module.device (torch.device) – Device on which to perform the computations. Default:
torch.device("cpu").g_first (bool) – whether to perform the step on \(g\) before that on \(f\) before or not. default: False
kwargs – Keyword arguments to be passed to the
deepinv.optim.BaseOptimclass.
- forward(y, physics, x_gt=None, compute_metrics=False)[source]
Runs the fixed-point iteration algorithm. This is the same forward as in the parent BaseOptim class, but without the
torch.no_grad()context manager.- Parameters:
y (torch.Tensor) – measurement vector.
physics (deepinv.physics) – physics of the problem for the acquisition of
y.x_gt (torch.Tensor) – (optional) ground truth image, for plotting the PSNR across optim iterations.
compute_metrics (bool) – whether to compute the metrics or not. Default:
False.
- Returns:
If
compute_metricsisFalse, returns (torch.Tensor) the output of the algorithm. Else, returns (torch.Tensor, dict) the output of the algorithm and the metrics.
Examples using BaseUnfold:
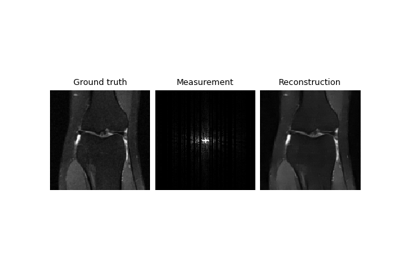
Self-supervised learning with Equivariant Imaging for MRI.
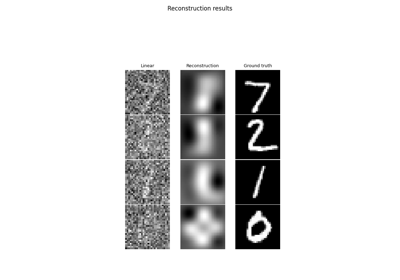
Learned Iterative Soft-Thresholding Algorithm (LISTA) for compressed sensing
Unfolded Chambolle-Pock for constrained image inpainting