Note
New to DeepInverse? Get started with the basics with the 5 minute quickstart tutorial..
Self-supervised learning with measurement splitting#
We demonstrate self-supervised learning with measurement splitting, to train a denoiser network on the MNIST dataset. The physics here is noisy computed tomography, as is the case in Noise2Inverse Hendriksen et al.[1]. Note this example can also be easily applied to undersampled multicoil MRI as is the case in SSDU Yaman et al.[2].
Measurement splitting constructs a ground-truth free loss \(\frac{m}{m_2}\| y_2 - A_2 \inversef{y_1}{A_1}\|^2\) by splitting the measurement and the forward operator using a randomly generated mask.
See deepinv.loss.SplittingLoss for full details.
from pathlib import Path
import torch
from torch.utils.data import DataLoader
from torchvision import transforms, datasets
import deepinv as dinv
from deepinv.utils import get_data_home
from deepinv.models.utils import get_weights_url
torch.manual_seed(0)
device = dinv.utils.get_device()
BASE_DIR = Path(".")
DATA_DIR = BASE_DIR / "measurements"
ORIGINAL_DATA_HOME = get_data_home()
Selected GPU 0 with 1763.25 MiB free memory
Define loss#
Our implementation has multiple optional parameters that control how the splitting is to be achieved. For example, you can:
Use
split_ratioto set the ratio of pixels used in the forward pass vs the loss;Define custom masking methods using a
mask_generatorsuch asdeepinv.physics.generator.BernoulliSplittingMaskGeneratorordeepinv.physics.generator.GaussianSplittingMaskGenerator;Use
eval_n_samplesto set how many realisations of the random mask is used at evaluation time;Optionally disable measurement splitting at evaluation time using
eval_split_input(as is the case in SSDU Yaman et al.[2]).Average over both input and output masks at evaluation time using
eval_split_output. Seedeepinv.loss.SplittingLossfor details.
Note that after the model has been defined, the loss must also “adapt” the model.
loss = dinv.loss.SplittingLoss(split_ratio=0.6, eval_split_input=True, eval_n_samples=5)
Prepare data#
We use the torchvision MNIST dataset, and use noisy tomography
physics (with number of angles equal to the image size) for the forward
operator.
Note
We use a subset of the whole training set to reduce the computational load of the example.
We recommend to use the whole set by setting train_datapoints=test_datapoints=None to get the best results.
transform = transforms.Compose([transforms.ToTensor()])
train_dataset = datasets.MNIST(
root=ORIGINAL_DATA_HOME, train=True, transform=transform, download=True
)
test_dataset = datasets.MNIST(
root=ORIGINAL_DATA_HOME, train=False, transform=transform, download=True
)
physics = dinv.physics.Tomography(
angles=28,
img_width=28,
noise_model=dinv.physics.noise.GaussianNoise(0.1),
device=device,
)
deepinv_datasets_path = dinv.datasets.generate_dataset(
train_dataset=train_dataset,
test_dataset=test_dataset,
physics=physics,
device=device,
save_dir=DATA_DIR,
train_datapoints=100,
test_datapoints=10,
)
train_dataset = dinv.datasets.HDF5Dataset(path=deepinv_datasets_path, train=True)
test_dataset = dinv.datasets.HDF5Dataset(path=deepinv_datasets_path, train=False)
train_dataloader = DataLoader(train_dataset, shuffle=True)
test_dataloader = DataLoader(test_dataset, shuffle=False)
/local/jtachell/deepinv/deepinv/deepinv/physics/tomography.py:188: UserWarning: The default value of `normalize` is not specified and will be automatically set to `True`. Set `normalize` explicitly to `True` or `False` to avoid this warning.
warn(
Dataset has been saved at measurements/dinv_dataset0.h5
Define model#
We use a simple U-Net architecture with 2 scales as the denoiser network.
To reduce training time, we use a pretrained model. Here we demonstrate training with 100 images for 1 epoch, after having loaded a pretrained model trained that was with 1000 images for 20 epochs.
Note
When using the splitting loss, the model must be “adapted” by the loss, as its forward pass takes only a subset of the pixels, not the full image.
model = dinv.models.ArtifactRemoval(
dinv.models.UNet(in_channels=1, out_channels=1, scales=2).to(device), pinv=True
)
model = loss.adapt_model(model)
optimizer = torch.optim.Adam(model.parameters(), lr=1e-3, weight_decay=1e-8)
# Load pretrained model
file_name = "demo_measplit_mnist_tomography.pth"
url = get_weights_url(model_name="measplit", file_name=file_name)
ckpt = torch.hub.load_state_dict_from_url(
url, map_location=lambda storage, loc: storage, file_name=file_name
)
model.load_state_dict(ckpt["state_dict"])
optimizer.load_state_dict(ckpt["optimizer"])
Downloading: "https://huggingface.co/deepinv/measplit/resolve/main/demo_measplit_mnist_tomography.pth?download=true" to /local/jtachell/.cache/torch/hub/checkpoints/demo_measplit_mnist_tomography.pth
0%| | 0.00/5.13M [00:00<?, ?B/s]
100%|██████████| 5.13M/5.13M [00:00<00:00, 66.4MB/s]
Train and test network#
To simulate a realistic self-supervised learning scenario, we do not use any supervised metrics for training, such as PSNR or SSIM, which require clean ground truth images.
Tip
We can use the same self-supervised loss for evaluation, as it does not require clean images,
to monitor the training process (e.g. for early stopping). This is done automatically when metrics=None and early_stop>0 in the trainer.
trainer = dinv.Trainer(
model=model,
physics=physics,
epochs=1,
losses=loss,
optimizer=optimizer,
device=device,
train_dataloader=train_dataloader,
eval_dataloader=test_dataloader,
metrics=None, # no supervised metrics
early_stop=2, # we can use early stopping as we have a validation loss
compute_eval_losses=True, # use self-supervised loss for evaluation
early_stop_on_losses=True, # stop using self-supervised eval loss
plot_images=False,
save_path=None,
verbose=True,
show_progress_bar=False,
no_learning_method="A_dagger", # use pseudo-inverse as no-learning baseline
)
model = trainer.train()
/local/jtachell/deepinv/deepinv/deepinv/training/trainer.py:1354: UserWarning: non_blocking_transfers=True but DataLoader.pin_memory=False; set pin_memory=True to overlap host-device copies with compute.
self.setup_train()
The model has 444737 trainable parameters
/local/jtachell/deepinv/deepinv/deepinv/loss/measplit.py:275: UserWarning: Mask generator not defined. Using new Bernoulli mask generator with shape torch.Size([40, 28]).
warn(
/local/jtachell/deepinv/deepinv/deepinv/physics/forward.py:932: UserWarning: At least one input physics is a DecomposablePhysics, but resulting physics will not be decomposable. `A_dagger` and `prox_l2` will fall back to approximate methods, which may impact performance.
warnings.warn(
Train epoch 0: TotalLoss=0.034
Eval epoch 0: TotalLoss=0.015
Best model saved at epoch 1
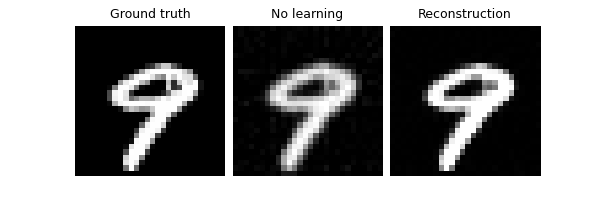
Test and visualize the model outputs using a small test set. We set the output to average over 5 iterations of random mask realisations. The trained model improves on the no-learning reconstruction by ~7dB.
trainer.plot_images = True
trainer.test(test_dataloader, metrics=dinv.metric.PSNR())
/local/jtachell/deepinv/deepinv/deepinv/training/trainer.py:1546: UserWarning: non_blocking_transfers=True but DataLoader.pin_memory=False; set pin_memory=True to overlap host-device copies with compute.
self.setup_train(train=False)
Eval epoch 0: TotalLoss=0.015, PSNR=6.444, PSNR no learning=-7.515
Test results:
PSNR no learning: -7.515 +- 0.386
PSNR: 6.444 +- 0.622
{'PSNR no learning': -7.514561700820923, 'PSNR no learning_std': 0.3862814005486887, 'PSNR': 6.444131755828858, 'PSNR_std': 0.6224634355378302}
Demonstrate the effect of not averaging over multiple realisations of
the splitting mask at evaluation time, by setting eval_n_samples=1.
We have a worse performance:
model.eval_n_samples = 1
trainer.test(test_dataloader, metrics=dinv.metric.PSNR())
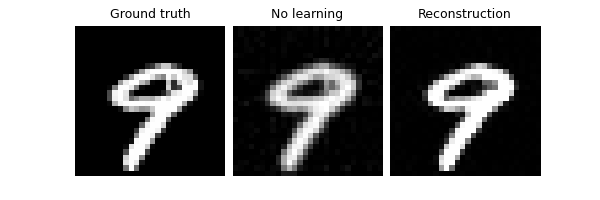
Eval epoch 0: TotalLoss=0.015, PSNR=2.954, PSNR no learning=-7.523
Test results:
PSNR no learning: -7.523 +- 0.364
PSNR: 2.954 +- 0.689
{'PSNR no learning': -7.5225166320800785, 'PSNR no learning_std': 0.3639746476424491, 'PSNR': 2.9536240816116335, 'PSNR_std': 0.6890917820921547}
Furthermore, we can disable measurement splitting at evaluation
altogether by setting eval_split_input to False (this is done in
SSDU Yaman et al.[2]). This generally is
worse than MC averaging:
model.eval_split_input = False
trainer.test(test_dataloader, metrics=dinv.metric.PSNR())
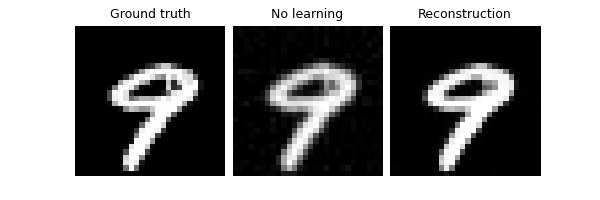
Eval epoch 0: TotalLoss=0.015, PSNR=0.312, PSNR no learning=-7.529
Test results:
PSNR no learning: -7.529 +- 0.389
PSNR: 0.312 +- 0.539
{'PSNR no learning': -7.528836297988891, 'PSNR no learning_std': 0.3888341828679185, 'PSNR': 0.3124205131083727, 'PSNR_std': 0.5392650487972105}
- References:
Total running time of the script: (0 minutes 22.979 seconds)

