SplittingLoss#
- class deepinv.loss.SplittingLoss(metric=None, split_ratio=0.9, mask_generator=None, eval_n_samples=5, eval_split_input=True, eval_split_output=False, pixelwise=True, normalize_loss=True)[source]#
Bases:
LossMeasurement splitting loss.
Implements measurement splitting loss. Splits the measurement and forward operator \(A\) (of size \(m\)) into two smaller pairs \((y_1,A_1)\) (of size \(m_1\)) and \((y_2,A_2)\) (of size \(m_2\)) , to compute the self-supervised loss:
\[\frac{m}{m_2}\| y_2 - A_2 \inversef{y_1}{A_1}\|^2\]where \(R\) is the trainable network, \(A_1 = M_1 A, A_2 = M_2 A\), and \(M_i\) are randomly generated masks (i.e. diagonal matrices) such that \(M_1+M_2=\mathbb{I}_m\).
See Self-supervised learning with measurement splitting for usage example.
Note
If the forward operator has its own subsampling mask \(M_{A}\), e.g.
deepinv.physics.Inpaintingordeepinv.physics.MRI, the splitting masks will be subsets of the physics’ mask such that \(M_1+M_2=M_{A}\)This loss was used for MRI in SSDU Yaman et al.[1] for MRI, Noise2Inverse Hendriksen et al.[2] for CT, as well as numerous other papers. Note we implement the multi-mask strategy proposed by Yaman et al.[1].
By default, the error is computed using the MSE metric, however any appropriate metric can be used.
Warning
The model should be adapted before training using the method
adapt_modelto include the splitting mechanism at the input.Note
To obtain the best test performance, the trained model should be averaged at test time over multiple realizations of the splitting, i.e. \(\hat{x} = \frac{1}{N}\sum_{i=1}^N \inversef{y_1^{(i)}}{A_1^{(i)}}\). To disable this, set
eval_n_samples=1.Note
To disable measurement splitting (and use the full input) at evaluation time, set
eval_split_input=False. This is done in SSDU Yaman et al.[1].Note
This loss allows training over images of varying shapes.
See also
deepinv.loss.mri.Artifact2ArtifactLoss,deepinv.loss.mri.Phase2PhaseLoss,deepinv.loss.mri.WeightedSplittingLoss,deepinv.loss.mri.RobustSplittingLossSpecialized splitting losses and their extensions for MRI applications.
- Parameters:
metric (Metric, torch.nn.Module) – metric used for computing data consistency, which is set as the mean squared error by default.
split_ratio (float) – splitting ratio, should be between 0 and 1. The size of \(y_1\) increases with the splitting ratio. Ignored if
mask_generatorpassed.mask_generator (deepinv.physics.generator.BernoulliSplittingMaskGenerator, None) – function to generate the mask. If None, the
deepinv.physics.generator.BernoulliSplittingMaskGeneratoris used, with the parameterssplit_ratioandpixelwise.eval_n_samples (int) – Number of samples used for averaging at evaluation time. Must be greater than 0.
eval_split_input (bool) – if True, perform input measurement splitting during evaluation. If False, use full measurement at eval (no MC samples are performed and eval_split_output will have no effect)
eval_split_output (bool) – at evaluation time, pass the output through the output mask too. i.e. \((\sum_{j=1}^N M_2^{(j)})^{-1} \sum_{i=1}^N M_2^{(i)} \inversef{y_1^{(i)}}{A_1^{(i)}}\). Only valid when \(y\) is same domain (and dimension) as \(x\). Although better results may be observed on small datasets, more samples must be used for bigger images. Defaults to
False.pixelwise (bool) – if
True, create pixelwise splitting masks i.e. zero all channels simultaneously. Ignored ifmask_generatorpassed.normalize_loss (bool) – whether to normalize loss by the target size
- Example:
>>> import torch >>> import deepinv as dinv >>> physics = dinv.physics.Inpainting(img_size=(1, 8, 8), mask=0.5) >>> model = dinv.models.MedianFilter() >>> loss = dinv.loss.SplittingLoss(split_ratio=0.9, eval_n_samples=2) >>> model = loss.adapt_model(model) # important step! >>> x = torch.ones((1, 1, 8, 8)) >>> y = physics(x) >>> x_net = model(y, physics, update_parameters=True) # save random mask in forward pass >>> l = loss(x_net, y, physics, model) >>> print(l.item() > 0) True
- References:
- class SplittingModel(model, split_ratio, mask_generator, eval_n_samples, eval_split_input, eval_split_output, pixelwise)[source]#
Bases:
ReconstructorModel wrapper when using SplittingLoss.
Performs input splitting during forward pass. At evaluation, perform forward passes for multiple realisations of splitting mask and average.
- Parameters:
model (deepinv.models.Reconstructor) – base model
split_ratio (float) – splitting ratio, should be between 0 and 1. The size of \(y_1\) increases with the splitting ratio. Ignored if
mask_generatorpassed.mask_generator (deepinv.physics.generator.PhysicsGenerator, None) – function to generate the mask. If None, the
deepinv.physics.generator.BernoulliSplittingMaskGeneratoris used, with the parameterssplit_ratioandpixelwise.eval_n_samples (int) – Number of samples used for averaging at evaluation time. Must be greater than 0.
eval_split_input (bool) – if True, perform input measurement splitting during evaluation. If False, use full measurement at eval (no MC samples are performed and eval_split_output will have no effect)
eval_split_output (bool) – at evaluation time, pass the output through the output mask too. i.e. \((\sum_{j=1}^N M_2^{(j)})^{-1} \sum_{i=1}^N M_2^{(i)} \inversef{y_1^{(i)}}{A_1^{(i)}}\). Only valid when \(y\) is same domain (and dimension) as \(x\). Although better results may be observed on small datasets, more samples must be used for bigger images. Defaults to
False.pixelwise (bool) – if
True, create pixelwise splitting masks i.e. zero all channels simultaneously. Ignored ifmask_generatorpassed.
- forward(y, physics, update_parameters=False)[source]#
Adapted model forward pass for input splitting. During training, only one splitting realisation is performed for computational efficiency.
- static split(mask, y, physics=None)[source]#
Perform splitting given mask
- Parameters:
mask (torch.Tensor) – splitting mask
y (torch.Tensor) – input data
physics (deepinv.physics.Physics) – physics to split, retaining its original noise model. If
None, only \(y\) is split.
- adapt_model(model, eval_n_samples=None)[source]#
Apply random splitting to input.
This method modifies a reconstruction model \(R\) to include the splitting mechanism at the input:
\[\hat{R}(y, A) = \frac{1}{N}\sum_{i=1}^N \inversef{y_1^{(i)}}{A_1^{(i)}}\]where \(N\geq 1\) is the number of Monte Carlo samples, and \(y_1^{(i)}\) and \(A_1^{(i)}\) are obtained by randomly splitting the measurements \(y\) and operator \(A\). During training (i.e. when
model.train()), we use only one sample, i.e. \(N=1\) for computational efficiency, whereas at test time, we use multiple samples for better performance. For other parameters that control how splitting is applied, see the class parameters.- Parameters:
model (torch.nn.Module) – Reconstruction model.
eval_n_samples (int) – deprecated. Pass
eval_n_samplesat class initialisation instead.
- Returns:
(
torch.nn.Module) Model modified for evaluation.- Return type:
- forward(x_net, y, physics, model, **kwargs)[source]#
Computes the measurement splitting loss
- Parameters:
x_net (torch.Tensor) – reconstructions.
y (torch.Tensor) – Measurements.
physics (deepinv.physics.Physics) – Forward operator associated with the measurements.
model (torch.nn.Module) – Reconstruction function.
- Returns:
(
torch.Tensor) loss.
- static split(mask, y, physics=None)[source]#
Perform splitting given mask
- Parameters:
mask (torch.Tensor) – splitting mask of shape (B,C,H,W)
y (torch.Tensor) – input data of shape (B,C,…,H,W)
physics (deepinv.physics.Physics) – physics to split, retaining its original noise model. If
None, only \(y\) is split.
Examples using SplittingLoss:#
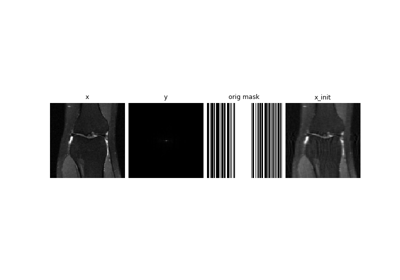
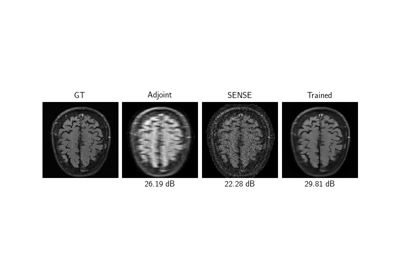
Self-supervised MRI reconstruction with Artifact2Artifact

Self-supervised learning with measurement splitting