FastMRISliceDataset#
- class deepinv.datasets.FastMRISliceDataset(root, target_root=None, load_metadata_from_cache=False, save_metadata_to_cache=False, metadata_cache_file='dataset_cache.pkl', slice_index='all', subsample_volumes=1.0, transform=None, filter_id=None, rng=None)[source]#
Bases:
ImageDataset,MRIMixinDataset for fastMRI that provides access to raw MR kspace data.
This dataset (from Knoll et al.[1]) randomly selects 2D slices from a dataset of 3D MRI volumes. This class considers one data sample as one slice of a MRI scan, thus slices of the same MRI scan are considered independently in the dataset.
To download raw data, please go to the bottom of the page
https://fastmri.med.nyu.edu/to download the brain/knee and train/validation/test volumes ash5files.The dataset is loaded as tuples
(x, y, params)where:yare the kspace measurements of shape(2, (N,) H, W)where N is the optional coil dimension depending on whether the data is singlecoil or multicoil. Note this kspace will be fully-sampled for training/validation datasets, and will be masked for test/challenge sets.x(“target”) are the (cropped) magnitude root-sum-square reconstructions of shape(1, H, W). If target is not present in the data (i.e. challenge/test set), thenxwill be returned astorch.nan. Optionally settarget_rootto load targets from a different directory.paramsis a dict containing parametersmaskand/orcoil_maps. Notemaskwill be automatically loaded if it is present (i.e. challenge/test set). Otherwise, you can generate masks and/or estimate coil maps usingdeepinv.datasets.fastmri.MRISliceTransform.
Tip
xandyare related bydeepinv.physics.MRI.A_adjoint()ordeepinv.physics.MultiCoilMRI.A_adjoint()depending on ifyare multicoil or not, withcrop=True, rss=True.Raw data file structure: (each file contains the k-space data and some metadata related to the scan)
self.root --- file1000005.h5 | -- xxxxxxxxxxx.h5.
When using this class, consider using the
metadata_cacheoptions to speed up class initialisation after the first initialisation.Note
We also provide a simple FastMRI dataset class in
deepinv.datasets.fastmri.SimpleFastMRISliceDataset. This allows you to save and load the dataset as 2D singlecoil slices much faster and all in-memory. You can generate this using the methodsave_simple_dataset.Important
By using this dataset, you confirm that you have agreed to and signed the FastMRI data use agreement.
See the fastMRI README for more details.
- Parameters:
root (str, pathlib.Path) – Path to the dataset.
target_root (str, pathlib.Path) – if specified, reads targets from files from this folder rather than root, assuming identical file structure. Defaults to None.
load_metadata_from_cache (bool) – Whether to load dataset metadata from cache.
save_metadata_to_cache (bool) – Whether to cache dataset metadata.
metadata_cache_file (str, pathlib.Path) – A file used to cache dataset information for faster load times.
subsample_volumes (float) – (optional) proportion of volumes to be randomly subsampled (float between 0 and 1).
slice_index (str, int, tuple) – if
"all", keep all slices per volume, ifint, keep only that indexed slice per volume, ifintortuple[int], index those slices, if"middle", keep the middle slice, if"middle+i", keep \(2i+1\) about middle slice, if"random", select random slice. Defaults to"all".transform (Callable) – optional transform function taking in (multicoil) kspace of shape (2, (N,) H, W) and targets of shape (1, H, W). Defaults to
deepinv.datasets.fastmri.MRISliceTransform.
See also
deepinv.datasets.MRISliceTransformTransform for working with raw data: simulate masks and estimate coil maps.
- Parameters:
filter_id (Callable) – optional function that takes
SliceSampleIDnamed tuple and returns whether this id should be included.rng (torch.Generator, None) – optional torch random generator for shuffle slice indices
For examples using raw data, see Tour of MRI functionality in DeepInverse.
- Examples:
Instantiate dataset with sample data (from a demo multicoil brain volume):
>>> from deepinv.datasets import FastMRISliceDataset, download_archive >>> from deepinv.utils import get_image_url, get_data_home >>> url = get_image_url("demo_fastmri_brain_multicoil.h5") >>> root = get_data_home() / "fastmri" / "brain" >>> download_archive(url, root / "demo.h5") >>> dataset = FastMRISliceDataset(root=root, slice_index="all") >>> len(dataset) 16 >>> target, kspace = dataset[0] >>> target.shape # (1, W, W), varies per sample torch.Size([1, 213, 213]) >>> kspace.shape # (2, N, H, W), varies per sample torch.Size([2, 4, 512, 213])
Load one slice per volume:
>>> dataset = FastMRISliceDataset(root=root, slice_index=0)
Use MRI transform to mask, estimate sensitivity maps, normalize and/or crop:
>>> from deepinv.datasets import MRISliceTransform >>> from deepinv.physics.generator import GaussianMaskGenerator >>> mask_generator = GaussianMaskGenerator((512, 213)) >>> dataset = FastMRISliceDataset(root, transform=MRISliceTransform(mask_generator=mask_generator, estimate_coil_maps=True)) >>> target, kspace, params = dataset[0] >>> params["mask"].shape torch.Size([1, 512, 213]) >>> params["coil_maps"].shape torch.Size([4, 512, 213])
Filter by volume ID:
>>> dataset = FastMRISliceDataset(root, filter_id=lambda s: "brain" in str(s.fname)) >>> len(dataset) 16
Convert to a simple normalized padded in-memory slice dataset from the middle slices only:
>>> simple_set = FastMRISliceDataset(root=root, slice_index="middle").save_simple_dataset(root.parent / "simple_set.pt") >>> len(simple_set) 1
Instantiate dataset with metadata cache (speeds up subsequent instantiation):
>>> dataset = FastMRISliceDataset(root=root, load_metadata_from_cache=True, save_metadata_to_cache=True, metadata_cache_file=root.parent / "cache.pkl") Saving dataset cache to ... >>> import shutil; shutil.rmtree(root.parent)
- References:
- class SliceSampleID(fname, slice_ind, metadata)[source]#
Bases:
NamedTupleData structure containing ID and metadata of specific slices within MRI data files.
- metadata_cache_manager(root, samples)[source]#
Read/write metadata cache file for populating list of sample ids.
- Parameters:
root (Union[str, pathlib.Path]) – root dir to save to metadata cache
samples (Any) – iterable (list, dict etc.) for populating with samples to read/write to metadata cache
- Yield:
samples, either populated from metadata cache, or blank, to be yielded to be written to.
- save_simple_dataset(dataset_path, pad_to_size=(320, 320), to_complex=False)[source]#
Convert dataset to a 2D singlecoil dataset and save as pickle file.
This allows the dataset to be loaded in memory with
deepinv.datasets.fastmri.SimpleFastMRISliceDataset.- Example:
Load local brain dataset and convert to simple dataset
from deepinv.datasets import FastMRISliceDataset root = "/path/to/dataset/fastMRI/brain/multicoil_train" dataset = FastMRISliceDataset(root=root, slice_index="middle") subset = dataset.save_simple_dataset(root + "/fastmri_brain_singlecoil.pt")
- Parameters:
- Returns:
loaded SimpleFastMRISliceDataset
- Return type:
- static torch_shuffle(x, generator=None)[source]#
Shuffle list reproducibly using torch generator.
- Parameters:
x (list) – list to be shuffled
generator (torch.Generator) – torch Generator.
- Return list:
shuffled list
- Return type:
Examples using FastMRISliceDataset:#
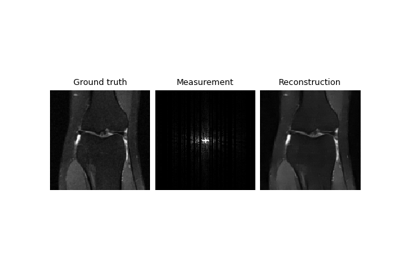
Self-supervised learning with Equivariant Imaging for MRI.